Trimeric SARS-CoV-2 Spike Proteins Produced from CHO Cells in Bioreactors Are High-Quality Antigens (Summary)
Author's note
In the early months of 2020, it became clear that the world was facing a new threat: a contagious, potentially lethal corona virus originated in Wuhan, China. This virus was named SARS-CoV-2 and it brought a new disease upon the world: COVID-19.
In January, Chinese scientists had released a full sequence of the viral RNA. By late February, the health situation around the world was serious, and Maria Wurm, CEO of ExcellGene, called upon her team. She wanted them to use their expertise to produce a part of the virus, to serve as a vaccine or diagnostic ingredient.
Scientists at ExcellGene had already faced a similar challenge in the years 2016-2019. Work had been carried out to produce a so-called “subunit vaccine” against Ebola virus. A subunit vaccine contains a small part of the original virus – in this case the GP1/GP2 proteins found on the surface of the Ebola virus. This exciting work, funded by EU Horizon 2020 and conducted in collaboration with 13 other research groups, led to the construction of tens of variants of the GP1 and GP2 proteins. Out of all the potential variants, one was chosen for full development.
Using a cell line from Chinese Hamster Ovary (CHO) cells, the scientists were able to produce the desired proteins in the form of a trimer – a complex structure consisting of three identical protein molecules. Subsequently the scientists effectively isolated and purified the protein trimers, which made them suitable for use in a vaccine. Results showed that merely one injection of the protein trimer was enough to give sterilizing immunity in mice, thereby stopping the virus from replicating entirely.
In May 2020, scientists at ExcellGene succeeded in making the same chinese hamster ovary cell line produce the Spike protein from the surface of SARS-CoV-2. In June, the production was so effective that tens of milligrams of high quality, highly purified protein could be delivered to research teams in EU and US. The preliminary studies of these proteins leave promising results and hopes for a valuable product.
The following will briefly provide key findings of the paper. Recently, after the publication, we found that the produced Spike protein induces a large production of antibodies by the immune systems of rabbits and horses. These levels of antibodies are enough to prevent infection in a cell culture model (Vero cells) of coronavirus infections.

Florian M. Wurm PhD, Prof. Emeri.
Chief Scientific Officer, Founder
Paper abstract
The Spike protein of SARS-CoV-2 is essential for virus entry into human cells. Most neutralizing antibodies against SARS-CoV-2 are directed against the Spike, making it the antigen of choice for use in vaccines and diagnostic tests. In the current pandemic context, global demand for Spike proteins has rapidly increased and could exceed hundreds of grams annually. Coronavirus Spikes are large, heavily glycosylated, homo-trimeric complexes, with inherent instability. The poor manufacturability now threatens availability of these proteins for vaccines and diagnostic tests. Here, we outline scalable, GMP-compliant, chemically defined processes for production of two cell secreted, stabilized forms of the trimeric Spike protein (Wuhan and D614G variants). The processes are chemically defined and based on clonal, suspension-CHO cell populations and on protein purification via a two-step, scalable down-stream process. The trimeric conformation was confirmed using electron microscopy and HPLC analysis. Binding to susceptible cells was shown using a virus-inhibition assay. The diagnostic sensitivity and specificity for detection of serum SARS-CoV-2 specific IgG1 was investigated and found to exceed that of Spike fragments (S1 and RBD). The process described here will enable production of sufficient high-quality trimeric Spike protein to meet the global demand for SARS-CoV-2 diagnostic tests and potentially vaccines.
Keywords: SARS-CoV-2; trimeric Spike; CHO cells; manufacturability; vaccines; diagnostics.
Paper details
Authors
Paco Pino , Joeri Kint , Divor Kiseljak, Valentina Agnolon , Giampietro Corradin , Andrey V. Kajava , Paolo Rovero, Ronald Dijkman, Gerco den Hartog , Jason S. McLellan, Patrick O. Byrne, Maria J. Wurm and Florian M. Wurm
Published in
Processes, 2020, 8(12), 1539; doi.org/10.3390/pr8121539
Introduction, synopsis of results and conclusions
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the virus responsible for the 2019 coronavirus disease (COVID-19) pandemic which presents an unprecedented challenge to societies globally. Effective vaccines and sensitive diagnostic tools for COVID-19 are urgently needed, and systems to produce and deliver these tools in sufficient quantities are required. As the SARS-CoV-2 trimeric Spike complex is a major target of the immune system, it could be a useful ingredient for vaccine and diagnostic applications. Stabilized trimeric Spike proteins have been selected as antigen of choice for RNA and virus-vector based vaccine candidates that are currently under development (Moderna, Novavax, Pfizer, J&J etc.). The efficacy of these vaccines was recently reported to be as high as 95 %. However, to maintain long-term protective immunity, follow-up vaccinations may be required. An adjuvated, subunit vaccine based on the stabilized trimeric Spike protein could be ideal for this purpose. Subunit vaccines can be produced in a cost-effective way and can be transported and stored lyophilized at ambient temperature. Also, whether any vaccination approach has elicited a sufficiently protective immune response against SARS-CoV-2 and the perpetuity of the immune response needs to be verified. For this reason, there will be a high demand for diagnostic tests for quantification of SARS-CoV-2-specific antibodies. The antigen which provides the best sensitivity and specificity for detection of SARS-CoV-2 antibodies is the trimeric form of the Spike protein a conclusion enforced by here presented data.
The viral Spike (S) protein complex is a surface-exposed homo-trimeric structure that mediates entry into host cells. The Spike engages the cellular host receptor ACE2 and allows virus-host membrane fusion. The Spike complex is considered the primary target of neutralizing antibodies. The ideal diagnostic test for SARS-CoV-2 antibodies would detect all antibodies directed against the trimeric Spike protein complex. Production of such diagnostic tests requires the production of large quantities of highly purified S protein in its natural prefusion conformation.
In this study, we developed a Chinese Hamster Ovary (CHO) cell based production process that yields high-quality trimeric Spike protein. This protein can be used for diagnostic tests as well as for vaccine applications. We used the fully characterized and CMC-compliant CHOExpress™ cell host (ExcellGene SA), single use equipment, chemically defined media and additives and obtained high-quality product at high volumetric productivity. This process can be readily scaled up to produce kilograms of trimeric Spike protein, should the demand rise to such a level. We showed that the purified trimeric Spike is efficient in blocking virus entry into cells when provided in molar excess to cultures infected with SARS-CoV-2. We also demonstrated the highest thus far seen sensitivity and specificity of Spike proteins for detection of Covid-19 patient-derived sera. We have modeled the product based on EM- images with both the “Wuhan” strain, as well as the D614G variant of the spike protein.
Using an optimized CHO expression system, and a scalable, chemically defined production process, trimeric SARS CoV-2 Spike proteins with mammalian-type glycosylation could be provided. The protein is of high purity and trimeric. Functional analysis showed it to be efficient in blocking virus infectivity in an in-vitro model. In addition diagnostic performance of the trimeric Spike for SARS-CoV-2 specific IgG was shown to exceed that of RDB and monomeric S1 protein.
Results
Design and selection of CHO manufacturable SARS-CoV-2 trimeric spikes
In order to define the optimal construct design to produce a soluble trimeric Spike protein we evaluated the expression with various S protein-variants (Figure 1) by CHO transient expression. The transmembrane domain and the C terminal intracellular tail were removed and replaced by a T4 foldon DNA sequence with or without (GGGS)n linker and a 8xHis tag encoding DNA. The furin cleavage site RRAR is mutated to be non-functional. WT Spike sequences of both the Wuhan strain and the D614G strain and amino-acid changes K986P/V987P (“2P”) for locking the protein into a prefusion conformation were also used. In addition, a receptor binding domain truncation of the trimeric Spike was used (RBD-His). S proteins variants with a scrambled furin cleavage site were found better expressed. Mutation of the furin cleavage site seemed to increase trimer assembly and/or stability, suggesting that trimer assembly via T4 foldon occurs in the Golgi (Trans-Golgi network where furin is active). The same construct modifications were also applied for the D614G mutation in the Spike protein which has recently replaced to a large extent the Wuhan version.
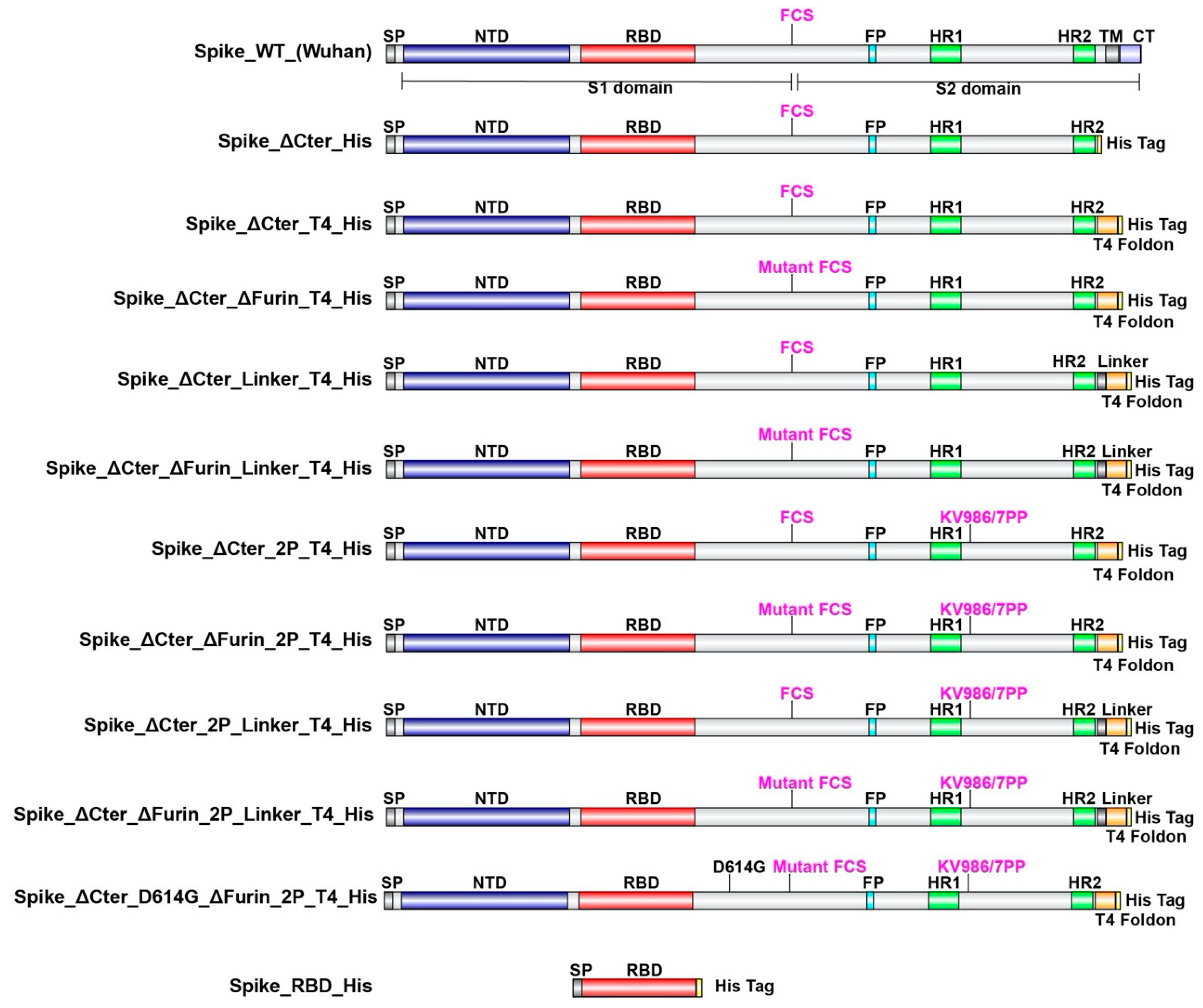
Schematic representation of SARS-CoV-2 spike protein designs. ΔCter refers to a construct that has the transmembrane and intracellular (TM, CT) section of the protein deleted. SP: Signal Peptide, NTD: N Terminal Domain, RBD: Receptor Binding Domain, FCS: Furin Cleavage Site, FP: Fusion Peptide, HR1: Heptad Repeat 1, HR2: Heptad Repeat 2, TM: Transmembrane Domain, CT: C terminal Tail.
The version named Spike_ΔCter_ΔFurin_2P_T4_His showed highest expression and appeared to be most promising in terms of trimeric assembly.
CHO expression and purification of SARS-CoV-2 proteins
Stable recombinant cell pools expressing trimeric Spike and RBD were generated using puromycin selection. From a total of 300 clonally derived cell populations, those with the highest expression levels of trimeric Spike and RBD were expanded and a fed-batch production process was developed. Optimal conditions were selected on the basis of product yield, product quality, viable cell density (VCD) and cell viability. The production process was scaled up to 200 mL shake flasks, to 10 L and to 40 L stirred tank bioreactors (STR). Viable cell density (VCD) and viability remained high for at least 10 days. From the authors’ experience in developing high-yielding manufacturing processes for clinically relevant recombinant antibodies and other proteins since almost 20 years, there is high confidence to assure feasibility to any scale of operation, up and beyond the 2,000 Liter scale.
Molecular characterization and electron microscopy analysis of trimeric Spikes
Trimeric Spike and RBD from cell-free culture supernatant were purified using an immobilized metal affinity chromatography (IMAC) capture step followed by preparative size exclusion. Eluates were concentrated and formulated at 1 mg/mL in PBS pH 7.4. Purity of the final products was estimated to be over 95 % as analyzed by HPLC-SEC (Figure 2 a and b).
HPLC-SEC (non-denaturing conditions) indicates that the S protein complex is trimeric (about 460 kDa) and the RBD is monomeric. The expected size of 150-160 kDa and 29 kDa for the Spike and RBD monomers, respectively, were confirmed by reducing SDS-PAGE (Figure 2 c, d). The heavy glycosylation of trimeric Spike contributes significantly to size heterogeneity, as has been seen with other CHO cell produced virus derived proteins, such as truncated surface proteins from Ebola and HIV. Trimeric confirmation was also confirmed using negative-stain electron microscopy and 3D modeling (Figure 2 d - g).
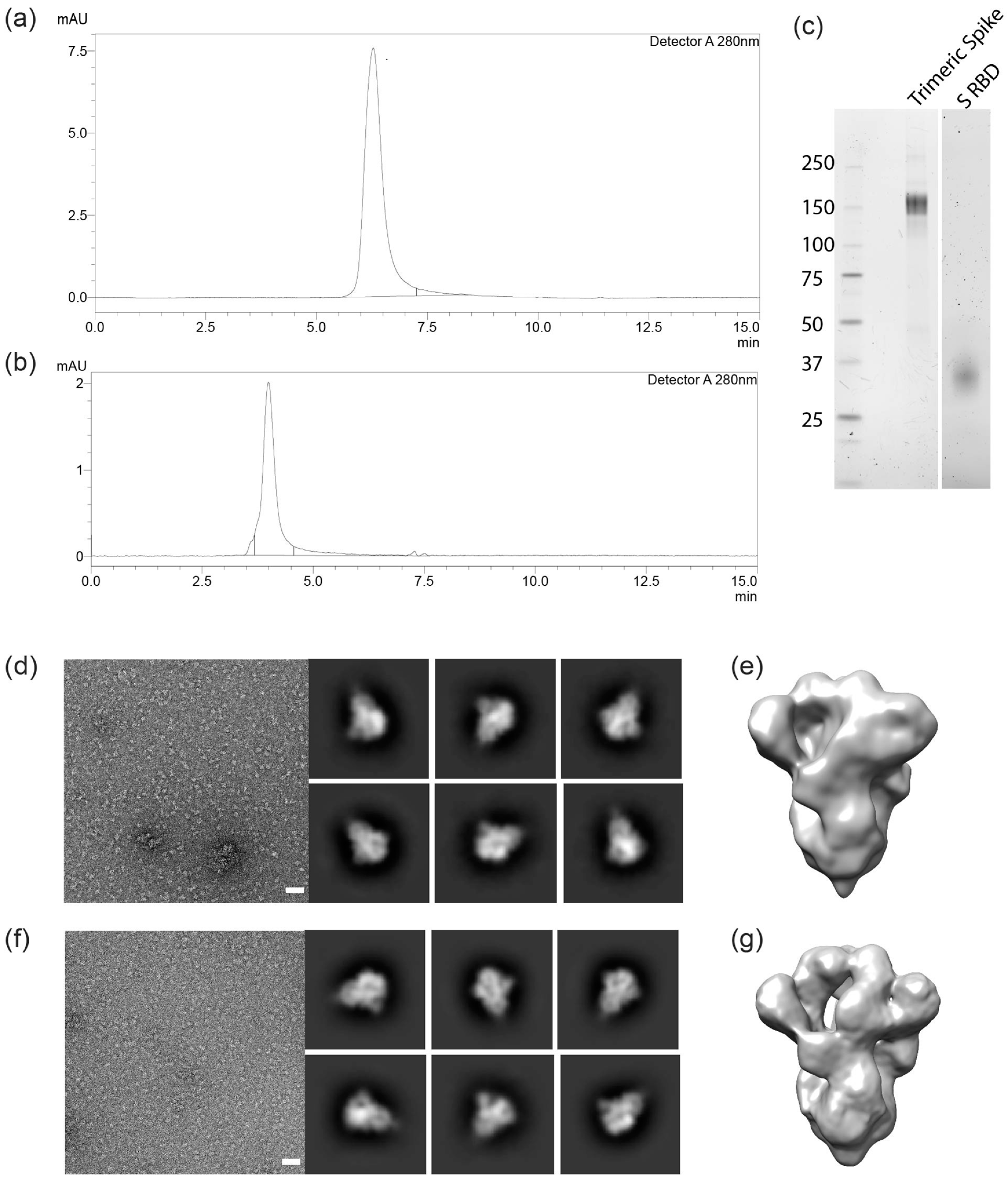
Characterization of the recombinant trimeric spike and RBD. (a,b): Size-exclusion chromatography (SEC) analysis plot with a single peak at 4.0 and 6.4 min for trimeric spike and RBD, respectively. SDS-PAGE gel showing bands for trimeric spike (c) and for RBD monomer (d). Negative-stain electron microscopy of the SARS-CoV-2 trimeric spike of the Wuhan strain (d,e) and for the D614G variant of the virus (f,g). White bars in d and f: 50 nm. Six 2-D class averages are shown to the right of each representative micrograph. 3-D reconstructions are shown in (e) and (g).
Inhibition of SARS-CoV-2 infection by SARS-CoV-2 proteins
SARS-CoV-2 virus was mixed with serially diluted RBD or trimeric Spike and the mixture was inoculated on Vero E6 cells. 48 hours later, SARS-CoV-2 infection was visualized and quantified (Figure 3 a and b). The presence of both the RBD Spike fragment and trimeric Spike reduced infection of Vero E6 cells. For RBD, reduction of infectivity was observed at a concentration of 20 µM and higher. For the trimeric Spike (both for the Wuhan variant and the D614G variant), a near 100-fold lower concentration reduced infection (Figure 3 b). Taken together, these results show that CHO-produced trimeric Spike and RBD are able to compete with SARS-CoV-2 for binding to host cells. No differences were observed between the 614D (Wuhan) and the D614G variants of the trimeric Spike.
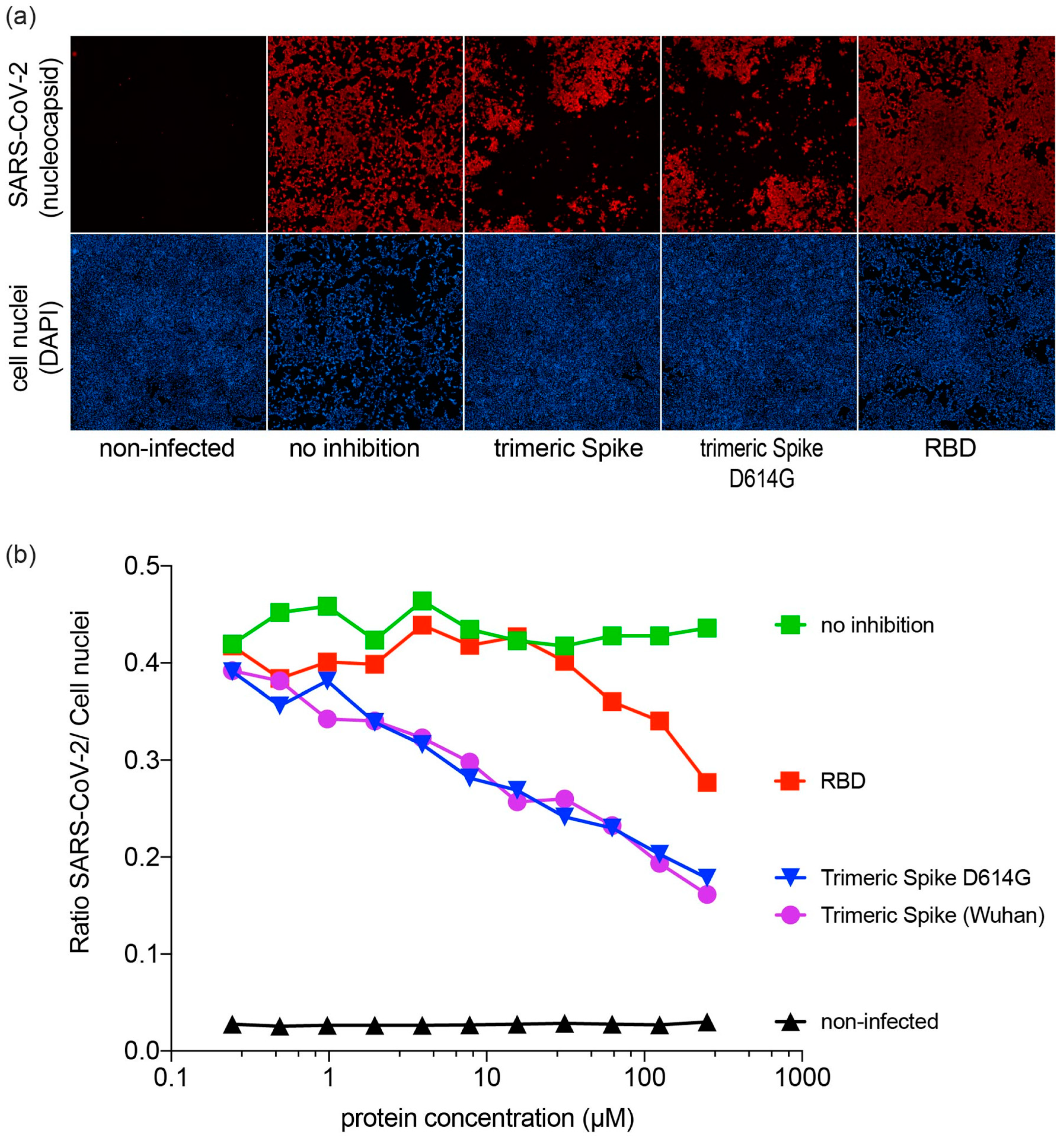
Inhibition of SARS-CoV-2 infection in Vero E6 cells; (a) Inhibitory effect of trimeric spike and RBD on infection of Vero E6 cells. SARS-CoV-2 antigen-positive cells were visualized by immunofluorescent staining. Virus nucleocapsid antigen staining (red), cell nucleus staining (blue); (b) Inhibitory effect of trimeric spike and RBD on infection of Vero E6 cells.
Sensitivity and specificity of SARS CoV-2 Spike proteins for detection of antibodies
The trimeric Spike and RBD proteins were used to detect SARS-CoV-2–specific IgG in a semi-quantitative bead-based serological assay. We specifically selected a set of COVID-19 serum samples that cover a large range of SARS-CoV-2–specific antibody concentrations. Sera (n = 72) were collected from PCR-confirmed non-hospitalized (n = 63) and hospitalized (n = 9) COVID-19 patients between days 4 and 40 after disease onset. Control sera (n = 79) were collected before the emergence of SARS-CoV-2 from patients with seasonal corona-induced Influenza-like illness (n = 44), non-corona, Influenza-like illness (n = 29), and from healthy individuals (n = 6).
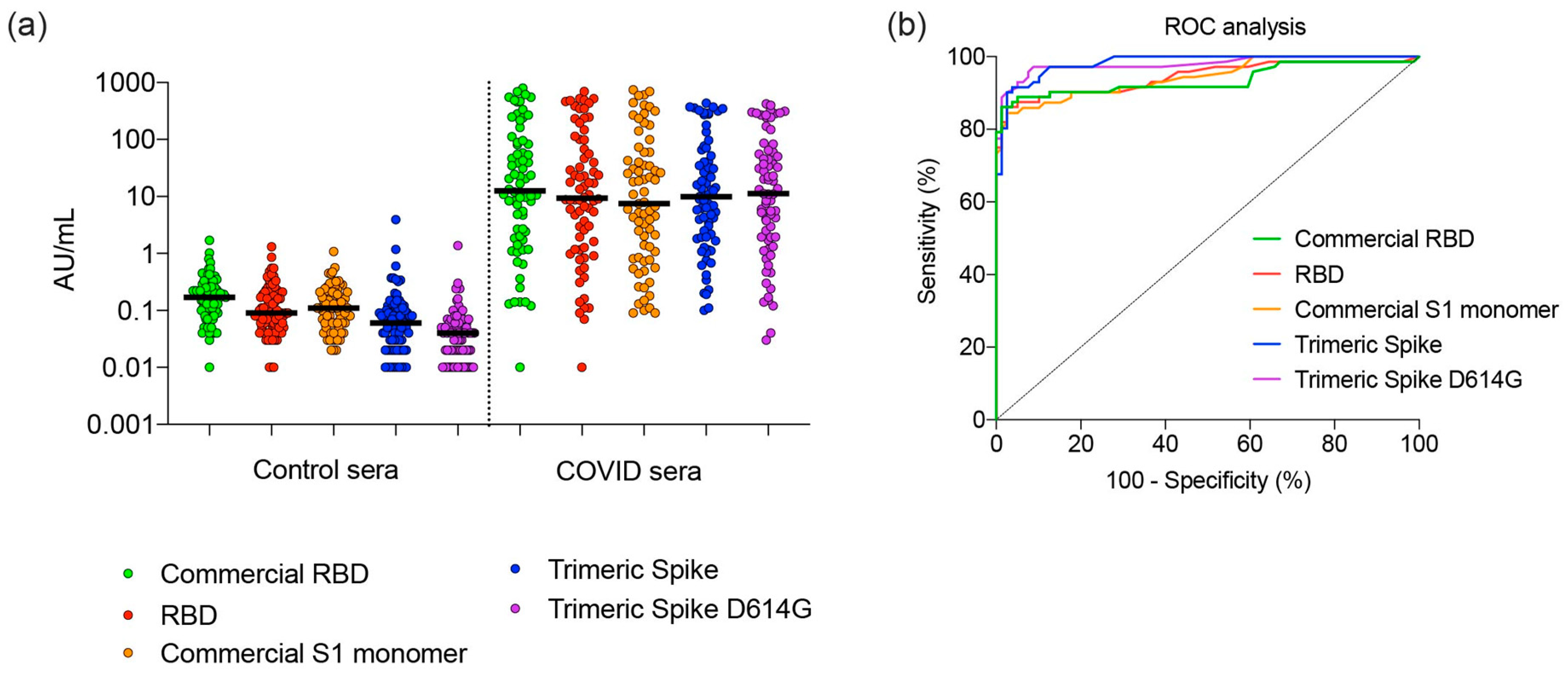
Performance of RBD, monomeric S1, and trimeric spike in a diagnostic assay to identify COVID-19 patients. (a) Control sera (n = 151) and COVID-19 sera (n = 72) collected at day 4–40 of symptoms were tested and compared for concentrations of immunoglobulin G. Median concentration and 95% confidence intervals are shown. (b) The sera tested in (a) were analyzed by ROC. Abbreviations: AU, arbitrary unit; RBD, receptor-binding domain; ROC, receiver operator characteristic; S1, Spike protein subunit 1.
We hope you enjoyed this shortened version of the paper. If you'd like to learn more, the full publication is available on the Processes website.
Interested in obtaining SARS-CoV-2 spike proteins?
Already know what you need? Make a request via the form below.